Wednesday, January 03, 2007
Molecular Anatomy of Influenza Virus Detailed
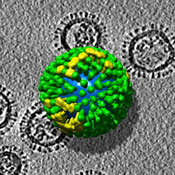
Scientists at the National Institute of Arthritis and Musculoskeletal and Skin Diseases** (NIAMS), part of the National Institutes of Health in Bethesda, Md., and colleagues at the University of Virginia in Charlottesville have succeeded in imaging, in unprecedented detail, the virus that causes influenza.
A team of researchers led by NIAMS' Alasdair Steven, Ph.D., working with a version of the seasonal H3N2 strain of influenza A virus, has been able to distinguish five different kinds of influenza virus particles in the same isolate (sample) and map the distribution of molecules in each of them. This breakthrough has the potential to identify particular features of highly virulent strains, and to provide insight into how antibodies inactivate the virus, and how viruses recognize susceptible cells and enter them in the act of infection.
"Being able to visualize influenza virus particles should boost our efforts to prepare for a possible pandemic flu attack," says NIAMS Director Stephen I. Katz, M.D., Ph.D. "This work will allow us to 'know our enemy' much better."
One of the difficulties that has hampered structural studies of influenza virus is that no two virus particles are the same. In this fundamental respect, it differs from other viruses; poliovirus, for example, has a coat that is identical in each virus particle, allowing it to be studied by crystallography.
The research team used electron tomography* (ET) to make its discovery. ET is a novel, three-dimensional imaging method based on the same principle as the well-known clinical imaging technique called computerized axial tomography, but it is performed in an electron microscope on a microminiaturized scale.
Original news release available via this link (Molecular Anatomy of Influenza Virus Detailed, December 29, 2006)
-------
Based on the Proceedings of the National Academy of Sciences (PNAS) paper:
Influenza virus pleiomorphy characterized by cryoelectron tomography
Audray Harris, Giovanni Cardone, Dennis C. Winkler, J. Bernard Heyman, Matthew Brecher, Judith M. White, and Alasdair C. Steven
PNAS | December 12, 2006 | vol. 103 | no. 50 | 19123-19127
Abstract
Influenza virus remains a global health threat, with millions of infections annually and the impending threat that a strain of avian influenza may develop into a human pandemic. Despite its importance as a pathogen, little is known about the virus structure, in part because of its intrinsic structural variability (pleiomorphy): the primary distinction is between spherical and elongated particles, but both vary in size. Pleiomorphy has thwarted structural analysis by image reconstruction of electron micrographs based on averaging many identical particles. In this study, we used cryoelectron tomography to visualize the 3D structures of 110 individual virions of the X-31 (H3N2) strain of influenza A. The tomograms distinguish two kinds of glycoprotein spikes [hemagglutinin (HA) and neuraminidase (NA)] in the viral envelope, resolve the matrix protein layer lining the envelope, and depict internal configurations of ribonucleoprotein (RNP) complexes. They also reveal the stems that link the glycoprotein ectodomains to the membrane and interactions among the glycoproteins, the matrix, and the RNPs that presumably control the budding of nascent virions from host cells. Five classes of virions, four spherical and one elongated, are distinguished by features of their matrix layer and RNP organization. Some virions have substantial gaps in their matrix layer ("molecular fontanels"), and others appear to lack a matrix layer entirely, suggesting the existence of an alternative budding pathway in which matrix protein is minimally involved.
-------
*A 2001 open access review from the Journal of Histochemistry and Cytochemistry:
The Emergence of Electron Tomography as an Important Tool for Investigating Cellular Ultrastructure
By Bruce F. McEwena and Michael Markoa
Journal of Histochemistry and Cytochemistry, Vol. 49, 553-564, May 2001
Abstract
Electron tomography has emerged as the leading method for the study of three-dimensional (3D) ultrastructure in the 5-20-nm resolution range. It is ideally suited for studying cell organelles, subcellular assemblies and, in some cases, whole cells. Tomography occupies a place in 3D biological electron microscopy between the work now being done at near-atomic resolution on isolated macromolecules or 2D protein arrays and traditional serial-section reconstructions of whole cells and tissue specimens. Tomography complements serial-section reconstruction by providing higher resolution in the depth dimension, whereas serial-section reconstruction is better able to trace continuity over long distances throughout the depth of a cell. The two techniques can be combined with good results for favorable specimens. Tomography also complements 3D macromolecular studies by offering sufficient resolution to locate the macromolecular complexes in their cellular context. The technology has matured to the point at which application of electron tomography to specimens in plastic sections is routine, and new developments to overcome limitations due to beam exposure and specimen geometry promise to further improve its capabilities. In this review we give a brief description of the methodology and a summary of the new insights gained in a few representative applications.
-------
** "The mission of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), a part of the Department of Health and Human Services’ National Institutes of Health, is to support research into the causes, treatment and prevention of arthritis and musculoskeletal and skin diseases; the training of basic and clinical scientists to carry out this research; and the dissemination of information on research progress in these diseases."
Technorati: national, institute, arthritis, skin, diseases, niams, imaging, influenza, virus, strain, a, cells, infection, pandemic, flu, research, electron, tomography, et, discovery, pnas, global, health, threat, human, 3-d, 2-d, technology, review, insights
Add to: CiteUlike | Connotea | Del.icio.us | Digg | Furl | Newsvine | Reddit | Yahoo